stat_tests – Statistical tests for datasets comparisons
Introduction
The available statistical tests are:
Student’s t-test, to compare two datasets, in the
studentmodule;The χ² test, for comparing distributions (spectra or meshes), in the
chi2module;The Bonferroni and Holm-Bonferroni tests, to solve the problem of multiple hypothesis tests, in the
bonferronimodule.
Modules
student — Student’s t-test
In Student’s t-test, we consider 2 different datasets that can have different means and different variances. The main problem is to test if the means are equal. One way to do that is to use Student’s t-test, although technically this name should be reserved for the case where the variances are unknown but they are known to be equal (the generalized version to different variances is called Welch’s test).
In Tripoli-4, like in most Monte Carlo codes, the estimated statistical uncertainty is the standard error, i.e. the standard deviation of the mean of the samples. It is typically assumed that the distribution of the mean is normal, as a consequence of the central limit theorem. The variance of the population is estimated as

and the variance of the mean is estimated as

The standard error  is the quantity that typically enters
Student’s t-test.
is the quantity that typically enters
Student’s t-test.
The Student test statistic is:

Note that, with our Dataset, this is
equivalent to
diff = dataset_1 - dataset_2
t(Student) = diff.value / diff.error
as value is difference of values and error is equal to the quadratic sum of the errors.
Test interpretation
The tested hypothesis in the Student’s t-test, i.e. the null hypothesis is:
equal means, so

Gaussian errors (or error following a normal distribution)
independent datasets.
The test interpretation is based on the comparison of the p-value with a given
significance level, called  . The (two-sided) p-value is defined
as the probability to observe results more extreme than those that were
actually observed, assuming that the null hypothesis is true. Mathematically,
. The (two-sided) p-value is defined
as the probability to observe results more extreme than those that were
actually observed, assuming that the null hypothesis is true. Mathematically,
 ,
,  being the Student
test statistic for samples of size
being the Student
test statistic for samples of size  ; the variable
; the variable  is also
called the number of degrees of freedom. The significativity level
is also
called the number of degrees of freedom. The significativity level
 is given by the user, usually 1% or 5%. Student’s test fails if
is given by the user, usually 1% or 5%. Student’s test fails if
 . If this is the case the
null hypothesis is rejected.
. If this is the case the
null hypothesis is rejected.
To evaluate the test we thus need an estimate of the number of degrees of
freedom, which is typically approximated as  , with
, with
 and
and  being the number of batches. For large values of
being the number of batches. For large values of
 (i.e. for a large number of degrees of freedom), the distribution
of the Student statistic approaches a standard normal distribution.
(i.e. for a large number of degrees of freedom), the distribution
of the Student statistic approaches a standard normal distribution.
It is sometimes easier to evaluate if the test succeeds by directly comparing
the value of  with a threshold given by the
significativity level
with a threshold given by the
significativity level  and the number of degrees of freedom.
and the number of degrees of freedom.
For example, here are some thresholds for some values of  and of
the number of degrees of freedom:
and of
the number of degrees of freedom:
|
|
|
N = 10 |
2.7638 |
1.8125 |
N = 1000 |
2.5807 |
1.9623 |
N = |
2.5758 |
1.9600 |
Results for N =  come from the normal distribution.
come from the normal distribution.
Note
This test can also be used on histograms (spectrum, mesh). In this case, an
independent test is made for each bin. The test for the whole histogram
succeeds if all the individual tests succeed. A better option is to use
a Bonferroni or a Holm-Bonferroni test (see the bonferroni
module).
Code implementation
If the number of degrees of freedom is given, p-values are estimated using Student’s t-distribution. If the number is not given or it is None, it is assumed that it is very large and the normal approximation is used.
The p-value is only calculated if the number of degrees of freedom is given;
otherwise, the Student result is just compared to the threshold. To test the
p-value instead of the value of the t-statistic, use
TestResultStudent.test_pvalue.
All these methods use the SciPy package (see scipy). The Student
distribution comes from scipy.stats.t, where the pdf is explicitely
written.
The method sf of scipy.stats.t corresponds to  , so
this gives the one-sided p-value; we need to multiply it by 2.0 to get the
two-sided p-value. The
, so
this gives the one-sided p-value; we need to multiply it by 2.0 to get the
two-sided p-value. The ppf (percent point) function returns the threshold
corresponding to the required one-sided significativity, so we need to divide
 by 2.0 in our case.
by 2.0 in our case.
The methods where the test is really applied are static methods, they can be
used outside of the TestStudent class. Examples are given in the class
docstrings.
Examples
Let’s get an example: one simulation gave 5.3 ± 0.2, the other one 5.25 ± 0.08. Are these numbers in agreement ?
>>> from valjean.eponine.dataset import Dataset
>>> from valjean.gavroche.stat_tests.student import TestStudent
>>> import numpy as np
>>> ds1 = Dataset(5.3, 0.2)
>>> ds2 = Dataset(5.25, 0.08)
>>> tstudent = TestStudent(ds1, ds2, name="comp",
... description="Comparison using Student's t-test")
>>> tstudent_res = tstudent.evaluate()
>>> print(f'{tstudent_res.tstud[0]:.7f}')
0.2321192
>>> print(f'{tstudent_res.test.threshold:.7f}')
2.5758293
>>> bool(tstudent_res)
True
To obtain the p-value, a number of degrees of freedom should be given:
>>> tstudent = TestStudent(ds1, ds2, ndf=1000, name="comp",
... description="Comparison using Student's t- test")
>>> tstudent_res = tstudent.evaluate()
>>> print(f'{tstudent_res.tstud[0]:.7f}')
0.2321192
>>> print(f'{tstudent_res.test.threshold:.7f}')
2.5807547
>>> bool(tstudent_res)
True
>>> print(f'{tstudent_res.pvalue[0]:.7f}')
0.8164929
>>> tstudent_res.test_pvalue()
[True]
This is also possible if we want to compare a bin to bin different spectra. Let’s define 2 small datasets containing arrays:
>>> ds3 = Dataset(np.array([5.2, 5.3, 5.25, 5.4, 5.5]),
... np.array([0.2, 0.25, 0.1, 0.2, 0.3]))
>>> ds4 = Dataset(np.array([5.1, 5.6, 5.2, 5.3, 5.2]),
... np.array([0.1, 0.3, 0.05, 0.4, 0.3]))
>>> tstudent = TestStudent(ds3, ds4, name="comp",
... description="Comparison using Student's t-test")
>>> tstudent_res = tstudent.evaluate()
>>> print(np.array2string(tstudent_res.oracles()))
[[ True True True True True]]
>>> bool(tstudent_res)
True
In the case of multiple comparisons, the test will return True if all the individual comparisons succeed. This is the case here.
It is possible to get a detailed bin-by-bin comparisons, including bad bins:
>>> ds5 = Dataset(np.array([5.1, 5.9, 5.8, 5.3, 4.5]),
... np.array([0.1, 0.1, 0.05, 0.4, 0.1]))
>>> tstudent = TestStudent(ds3, ds5, name="comp",
... description="Comparison using Student's t-test")
>>> tstudent_res = tstudent.evaluate()
>>> print(f'{tstudent_res.test.threshold:.7f}')
2.5758293
>>> print(np.array2string(tstudent_res.oracles()))
[[ True True False True False]]
Since some of the comparisons failed here, the whole tests fails as well:
>>> bool(tstudent_res)
False
As in the scalar case, it is possible to ask for the p-values. In both cases
the significativity can be changed via the alpha argument, as shown here:
>>> tstudent = TestStudent(ds3, ds5, name="comp", alpha=0.05, ndf=1000,
... description="Comparison using Student's t-test")
>>> tstudent_res = tstudent.evaluate()
>>> print(f'{tstudent_res.test.threshold:.7f}')
1.9623391
>>> print(np.array2string(tstudent_res.oracles()))
[[ True False False True False]]
>>> print(f'{tstudent_res.tstud[0][1]:.7f}')
-2.2283441
This last value is in-between the thresholds at 1% and 5%, in magnitude, and therefore the test succeeds in the former case and fails in the latter.
Obtaining p-values and comparing them is also possible in the spectrum case:
>>> print(np.array2string(tstudent_res.pvalue[0],
... formatter={'float_kind':'{:.7e}'.format}))
[6.5481769e-01 2.6079281e-02 1.0147751e-06 8.2310894e-01 1.6125524e-03]
>>> print(np.array2string(tstudent_res.test_pvalue()[0]))
[ True False False True False]
Finally, for an array of size 1 (and dimension 1), the test will do the same as
for datasets containing numpy.generic:
>>> ds6 = Dataset(np.array([5.3]), np.array([0.2]))
>>> ds7 = Dataset(np.array([5.25]), np.array([0.08]))
>>> tstudent = TestStudent(ds6, ds7, name="comp",
... description="Comparison using Student's t-test")
>>> tstudent_res = tstudent.evaluate()
>>> print(np.array2string(tstudent_res.tstud[0],
... formatter={'float_kind':'{:.7f}'.format}))
[0.2321192]
>>> print(f'{tstudent_res.test.threshold:.7f}')
2.5758293
>>> bool(tstudent_res)
True
Like for usual operations on datasets, only tests between datasets of the same shape are possible:
>>> tstudent = TestStudent(ds1, ds7, name="comp",
... description="Comparison using Student's t-test")
>>> tstudent_res = tstudent.evaluate()
Traceback (most recent call last):
...
ValueError: Datasets to subtract do not have same shape
Multi-dimensional datasets are also allowed:
>>> ds8 = Dataset(np.array([[5.2, 5.3, 5.25], [5.4, 5.5, 5.2]]),
... np.array([[0.2, 0.25, 0.1], [0.2, 0.3, 0.1]]))
>>> ds9 = Dataset(np.array([[5.1, 5.6, 5.2], [5.3, 5.2, 5.3]]),
... np.array([[0.1, 0.3, 0.05], [0.4, 0.3, 0.2]]))
>>> ds8.shape
(2, 3)
>>> ds8.size
6
>>> tstudent = TestStudent(ds8, ds9, alpha=0.05, ndf=1000, name="comp",
... description="Comparison using Student's t-test")
>>> tstudent_res = tstudent.evaluate()
>>> bool(tstudent_res)
True
>>> print(np.array2string(tstudent_res.tstud[0],
... formatter={'float_kind':'{:.7f}'.format}))
[[0.4472136 -0.7682213 0.4472136]
[0.2236068 0.7071068 -0.4472136]]
>>> print(np.array2string(tstudent_res.oracles()))
[[[ True True True]
[ True True True]]]
Warning
If the errors are equal to 0, so the Student denominator is 0, AND the values are equal, so the numerator is also 0, the Student value is set to 0. In that case the test is assumed to succeed (boolean returning True).
>>> ds10 = Dataset(np.array([3.2, 0, 5]), np.array([0, 0.5, 0]))
>>> ds11 = Dataset(np.array([3.2, 0, 5.2]), np.array([0, 0.5, 0]))
>>> tstudent = TestStudent(ds10, ds11, name='zeros')
>>> tstudent_res = tstudent.evaluate()
>>> bool(tstudent_res)
False
>>> print(np.array2string(tstudent_res.tstud[0]))
[ 0. 0. -inf]
>>> print(np.array2string(tstudent_res.oracles()))
[[ True True False]]
- class valjean.gavroche.stat_tests.student.TestResultStudent(test, tstud, pvalue=None)[source]
Result from Student’s t-test.
- __init__(test, tstud, pvalue=None)[source]
Initialisation of
TestResultStudentMembers are lists whose length corresponds to the number of datasets compared to the reference dataset.
- Parameters:
test (
TestStudent) – the TestStudent objecttstud (
list(numpy.ndarray)) – Student’s t-test resultspvalue (
list(numpy.generic) orlist(list(numpy.generic))) – p-value or p-values depending on datasets, default is None
- test_alpha(tstud)[source]
Test p-value or first kind error.
- Parameters:
tstud (
numpy.generic) – value of Student’s t-statistic to be used for comparison- Returns:
bool
- __bool__()[source]
Has this test succeeded?
The policy for
TestResultStudentis that the test is considered to fail if any of the p-values are below the given significance level. This means that a test on aDatasethaving multiple bins will fail if any of the bins exhibits large fluctuations. You might want to useTestBonferroniorTestHolmBonferronito deal with such cases.If this test concerns multiple datasets, the test is considered to succeed if all the dataset individually pass the test against the reference.
- test_pvalue()[source]
Result of the test by testing p-value.
- Returns:
bool or
numpy.ndarrayof bool ifndfwas given toTestStudentand datasets arenumpy.ndarray
- class valjean.gavroche.stat_tests.student.TestStudent(dsref, *datasets, name, description='', labels=None, alpha=0.01, ndf=None)[source]
Class to build the Student’s t-test.
- __init__(dsref, *datasets, name, description='', labels=None, alpha=0.01, ndf=None)[source]
Initialisation of
TestStudent- Parameters:
name (str) – local name of the test
description (str) – specific description of the test
labels (dict) – labels to be used for test classification in reports (for example category, input file name, type of result, …)
dsref (
Dataset) – reference datasetdatasets (
list(Dataset)) – list of datasets to be compared to reference datasetalpha (float) – significance level, i.e. limit on the p-value (expected values are typically 0.01, 0.05), default is
0.01ndf (int) – default is
None, if given p-value will be calculated for ndf degrees of freedom (should correspond to number of batches), otherwise ndf assumed infinite, normal approximation
- static student_test(ds1, ds2)[source]
Compute Student’s t-test or Student distribution for the given datasets (static method).
- Parameters:
- Returns:
numpy.genericornumpy.ndarraydepending onDatasettype
- static pvalue(tstud, ndf)[source]
Calculation of the p-value (static method).
- Parameters:
tstud (
numpy.generic(float) ornumpy.ndarray(float)) – Student’s t-test statisticndf (int or
numpy.generic(int)) – number of degrees of freedom
- Returns:
numpy.generic(float) ornumpy.ndarray(float)
- static student_threshold(alpha, ndf=None)[source]
Get threshold from probability (static method).
- Parameters:
alpha (float or
numpy.generic(float)) – required significance level ( )
)ndf (int or
numpy.generic(int), default isNone) – number of degrees of freedom
- Returns:
The threshold is two-sided.
If number of degrees of freedom is given, the distribution used is the Student distribution corresponding to
ndf. If not, we consider an infinite number of degrees of freedom. In that case, the Student distribution tends to a normal distribution.
chi2 – 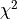 test
test
In our case we’ll use the modified χ²-test widely used in physics for comparing
histograms. This test is also available in the ROOT
software. For further explanations a good desciption
is provided by the documentation of ROOT’s Chi2Test() method on
TH1 page or in this
article by N. Gagunashvili.
The used formula is then:

where datasets (or distributions) 1 and 2 are compared,  being the
values and
being the
values and  being the error in each bin.
being the error in each bin.
In our case only the ‘weighted’ case is implemented.
Statistical interpretation
The null hypothesis for the χ²-test is:
equal means in each bin
gaussian means in each bin (so normal distribution in each bin)
bins are independent
The interpretation of the test is based on the χ² value and on the p-value. Usually it is expected that the χ²/ndf should be close to 1 (ndf = number of degrees of freedom), see PDG, statistics chapter. The p-value can also be calculated from the probability density function of the χ² distribution, which is given by:

for  ,
,  the number of degrees of freedom and
the number of degrees of freedom and
 is Euler’s Gamma function [1].
is Euler’s Gamma function [1].
The cumulative distribution function,  , used to calculated the
p-value is then:
, used to calculated the
p-value is then:

where  is the lower incomplete gamma function
[2].
is the lower incomplete gamma function
[2].
It is equivalent to calculate the probability  which
is equal to
which
is equal to  , so the integral from
, so the integral from  to
to
 . This probability is the p-value.
. This probability is the p-value.
If the p-value is lower than the define level (1%, 5% for example), the null hypothesis is rejected.
Possible biases can come from wrong estimation of uncertainties (underestimated for example or only statistical while a systematic one should be taken into account).
Code implementation
SciPy is used to calculate the p-value. The distribution
scipy.stats.chi2 is imported as schi2` in order to name conflicts. The
p-value is obtained with the sf method. It is directly compared with the
significance level required by the user.
Warning
If some of your bins are not filled, if they are initialized to zero or if
their error is zero, the χ² calculation will be wrong and probably give
 .
.
One option is proposed to remove these bins from the calculation,
ignore_empty. The use of this option implies a reduction of the number
of degrees of freedom by the number of removed bins (ndf = # bins). It has
to be considered with caution, this does not make necessarly sense,
depending on the user case.
Another option is to apply some rebinning in order to have no empty bins (what usually means re-running to get consistent uncertainties).
Examples of the test
Let’s consider a spectrum of 5 bins with their error and apply the χ²-test.
>>> from valjean.eponine.dataset import Dataset
>>> from valjean.gavroche.stat_tests.chi2 import TestChi2
>>> import numpy as np
>>> ds1 = Dataset(np.array([5.2, 5.3, 5.25, 5.4, 5.5]),
... np.array([0.2, 0.25, 0.1, 0.2, 0.3]))
>>> ds2 = Dataset(np.array([5.1, 5.6, 5.2, 5.3, 5.2]),
... np.array([0.1, 0.3, 0.05, 0.4, 0.3]))
>>> tchi2 = TestChi2(ds1, ds2, alpha=0.05, name="comp",
... description="Comparison using Chi2 test")
>>> tchi2_res = tchi2.evaluate()
>>> print(f'{tchi2_res.chi2_per_ndf[0]:.7f}')
0.3080328
>>> bool(tchi2_res)
True
>>> print(f'{tchi2_res.pvalue[0]:.7f}')
0.9083889
Test with empty bins
One “empty bin” at the same position in the 2 compared datasets prevents the
test to work. An empty bin is defined as a bin with zero error. In Monte
Carlo codes a zero error can happen for example when only one batch has been
run, no variance can be calculated in that case (n-1 usually used with
n the number of batches).
>>> ds3 = Dataset(np.array([5.2, 5.3, 5.25, 5.4, 5.5]),
... np.array([0.2, 0.25, 0., 0.2, 0.3]))
>>> ds4 = Dataset(np.array([5.1, 5.6, 5.2, 5.3, 5.2]),
... np.array([0.1, 0.3, 0., 0.4, 0.3]))
>>> tchi2 = TestChi2(ds3, ds4, alpha=0.05, name="comp",
... description="Comparison using Chi2 test")
>>> tchi2_res = tchi2.evaluate()
>>> tchi2_res.chi2
[inf]
>>> bool(tchi2_res)
False
>>> print(np.array2string(tchi2_res.test.nonzero_bins[0]))
[ True True True True True]
>>> tchi2_res.test.ndf[0]
5
>>> np.count_nonzero(ds4.error)
4
If the values are also at zero, we get a nan instead of an inf.
To get a χ² evaluation based on the non-empty bins, use the ignore_empty
argument of the TestChi2. It recalculates the number of degrees of
freedom removing the empty bins and only use the non-zero ones in the χ²
calculation.
>>> tchi2 = TestChi2(ds3, ds4, alpha=0.05, ignore_empty=True,
... name="comp", description="Comparison using Chi2 test")
>>> tchi2_res = tchi2.evaluate()
>>> print(f'{tchi2_res.chi2[0]:.7f}')
1.3401639
>>> tchi2_res.test.ndf[0]
4
>>> tchi2_res.test.dsref.size
5
>>> print(np.array2string(tchi2_res.test.nonzero_bins[0]))
[ True True False True True]
>>> print(f'{tchi2_res.chi2_per_ndf[0]:.7f}')
0.3350410
>>> bool(tchi2_res)
True
Test with multiple dimensions datasets
>>> ds5 = Dataset(np.array([[5.2, 5.3, 5.25], [5.4, 5.5, 5.2]]),
... np.array([[0.2, 0.25, 0.1], [0.2, 0.3, 0.1]]))
>>> ds6 = Dataset(np.array([[5.1, 5.6, 5.2], [5.3, 5.2, 5.3]]),
... np.array([[0.1, 0.3, 0.05], [0.4, 0.3, 0.2]]))
>>> ds5.shape
(2, 3)
>>> ds5.size
6
>>> tchi2 = TestChi2(ds5, ds6, alpha=0.05, name="comp",
... description="Comparison using Chi2 test")
>>> tchi2_res = tchi2.evaluate()
>>> print(f'{tchi2_res.chi2_per_ndf[0]:.7f}')
0.2900273
>>> bool(tchi2_res)
True
>>> print(f'{tchi2_res.pvalue[0]:.7f}')
0.9419786
And when we have at least one empty bin, using the ignore_empty argument:
>>> ds7 = Dataset(np.array([[5.2, 5.3, 5.25], [5.4, 5.5, 5.2]]),
... np.array([[0.2, 0.25, 0.], [0.2, 0.3, 0.1]]))
>>> ds8 = Dataset(np.array([[5.1, 5.6, 5.2], [5.3, 5.2, 5.4]]),
... np.array([[0.1, 0.3, 0.], [0.4, 0.3, 0.]]))
>>> ds7.shape
(2, 3)
>>> ds7.size
6
>>> tchi2 = TestChi2(ds7, ds8, alpha=0.05, ignore_empty=True,
... name="comp", description="Comparison using Chi2 test")
>>> tchi2_res = tchi2.evaluate()
>>> print(f'{tchi2_res.chi2[0]:.7f}')
5.3401639
>>> tchi2_res.test.ndf[0]
5
>>> tchi2_res.test.dsref.size
6
>>> print(np.array2string(tchi2_res.test.nonzero_bins[0]))
[[ True True False]
[ True True True]]
>>> print(f'{tchi2_res.chi2_per_ndf[0]:.7f}')
1.0680328
>>> bool(tchi2_res)
True
Note
A RuntimeWarning is emitted if zero bins are used during calculation.
Footnotes
- class valjean.gavroche.stat_tests.chi2.TestResultChi2(test, chi2, pvalue)[source]
Result from the χ²-test.
- __init__(test, chi2, pvalue)[source]
Construct a TestResultChi2 object.
Members are lists which length corresponds to the number of datasets compared to the reference dataset.
- Parameters:
test (
TestChi2) – the χ² test objectchi2 (
list(numpy.generic(float))) – the value of the χ² statisticpvalue (
list(numpy.generic(float))) – the test p-value
- class valjean.gavroche.stat_tests.chi2.TestChi2(dsref, *datasets, name, description='', labels=None, alpha=0.01, ignore_empty=False)[source]
Test class for χ², inheritate from
Test.- __init__(dsref, *datasets, name, description='', labels=None, alpha=0.01, ignore_empty=False)[source]
Initialisation of
TestChi2- Parameters:
name (str) – local name of the test
description (str) – specific description of the test
labels (dict) – labels to be used for test classification in reports (for example category, input file name, type of result, …)
dsref (
Dataset) – reference datasetdatasets (
list(Dataset)) – list of datasets to be compared to reference datasetalpha (float) – probability to accept of not the test (p-value is expected greater), i.e. significance level
ignore_empty (bool) – if the array contains zero bins, the test can done not consider them if this option is True; otherwise it will probably fail as the array to sum will contain infinite terms. Default is
False.
- nonzero_bins
nonzero bins identification by True, False if zero (
list(numpy.ndarray(bool)))
- static pvalue(chi2, ndf)[source]
Calculation of the p-value of the test.
- Parameters:
chi2 (
numpy.generic(float)) – observed χ²ndf (int or
numpy.generic(int)) – number of degrees of freedom
- Returns:
- static chi2_test(ds1, ds2, nonzero_bins=None)[source]
Compute the χ² value for the given datasets.
- Parameters:
ds1 (
Dataset) – first datasetds2 (
Dataset) – second datasetnonzero_bins (
numpy.ndarray(bool) orNone(default)) – optional argument, booleans array of the same size asds1andds2to identify zero bins and possibly avoid them (see below)
- Returns:
χ² value
- Return type:
It has to be noted that if in a bin the sum of the variances of the test and the reference dataset is zero (so both datasets bin has a zero error) the bin cannot be taken into account in the calculation of the sum as the ratio will be infinite. This is why they can be removed via the
nonzero_binsargument at that step. This has another consequence: the number of degrees of freedom is consequently reduced by the number of bins removed.
bonferroni – Multiple hypothesis tests
In some cases we would like to interpret not only one test (like on a generic score in Tripoli-4) but multiple ones (in a spectrum case for example). Each individual test is supposed to be successful but in most of the case we can accept that some of them fail (in a given acceptance of course).
The Bonferroni correction and the Holm-Bonferroni method allow to treat such cases. See for example family-wise error rate Wikipedia webpage for more details on this kind of tests.
Here are implemented two tests based on comparison of the p-value from the chosen test and a significance level:
Bonferroni correction: all individual p-values are compared to the same significance level, depending on the number of tests;
Holm-Bonferroni method: p-values are first sorted then compared to different significance levels depending on number of tests and rank.
In both cases a first test is chosen, like Student test, to evaluate the individual hypothesis. The p-value of the test is calculated and used in the following tests. Like in other tests, we consider two-sided significance levels here.
The null hypothesis is that the p-value is higher than a given significance level, that depends on the overall required significance level, the number of tests considered and the chosen method (Bonferroni or Holm-Bonferroni).
Bonferroni correction
The Bonferroni correction is used for multiple comparisons. It is based on a weighted comparison to the significance level α.
If we have  different tests (for example a spectrum with
different tests (for example a spectrum with  bins), the significance level α will be weighted by
bins), the significance level α will be weighted by  . This p-value of
the
. This p-value of
the  tests will then be compared to that value. As a consequence, the
null hypothesis will be rejected if
tests will then be compared to that value. As a consequence, the
null hypothesis will be rejected if

The individual significance level is usually largely smaller than the required one and the sum of them obvioulsy give α.
Conclusion of the test depends on what we want:
it can be a simple
Trueif all hypothesis are accepted;it can be
Falseif at least one null hypothesis was rejected;we can also choose a level of acceptance, for example “it is fine if 1 % of the null hypothesis are rejected”
Per default, in our case the test is rejected if there is at least one null hypothesis rejected.
Holm-Bonferroni method
The Holm-Bonferroni method is a variation of the Bonferroni correction.
The p-value from the chosen test are first sorted from lower to higher value.
Then each of them are compared to the significance level weighted by
 with
with  the number of tests and
the number of tests and  the rank
(starting at 1). The null hypothesis is rejected if
the rank
(starting at 1). The null hypothesis is rejected if

The test can be stopped at the first  accepting the null hypothesis.
accepting the null hypothesis.
This test is quite conservative. It can be possible to get the proportion of accepted and rejected hypothesis, like in the Bonferroni correction case.
Code implementation
In both cases, the test is initiliazed with another test, a Student test for
example. This test can be evaluated inside the new test
evaluate method or outside of it. Only examples with
the full structure will be shown here.
TestResults are returned as False if one null hypothesis is rejected.
Access to the initial test is still provided.
Examples
Let’s do the usual imports then use the quick example from Student test, with the p-value calculation:
>>> import numpy as np
>>> from valjean.eponine.dataset import Dataset
>>> from valjean.gavroche.stat_tests.student import TestStudent
>>> from valjean.gavroche.stat_tests.bonferroni import TestBonferroni
>>> from valjean.gavroche.stat_tests.bonferroni import TestHolmBonferroni
Successful test with Bonferroni correction and Holm-Bonferroni method
>>> ds1 = Dataset(np.array([5.2, 5.3, 5.25, 5.4, 5.5]),
... np.array([0.2, 0.25, 0.1, 0.2, 0.3]))
>>> ds2 = Dataset(np.array([5.1, 5.6, 5.2, 5.3, 5.2]),
... np.array([0.1, 0.3, 0.05, 0.4, 0.3]))
>>> tstudent_12 = TestStudent(ds1, ds2, name="comp",
... description="Comparison using Student test",
... alpha=0.05, ndf=1000)
>>> tbonf = TestBonferroni(name="bonf", description="Bonferroni correction",
... test=tstudent_12, alpha=0.05)
>>> tbonf.ntests
5
>>> tbonf.ntests == ds1.size
True
>>> tbonf.bonf_signi_level
0.005
>>> tbonf_res = tbonf.evaluate()
>>> bool(tbonf_res)
True
>>> tbonf_res.nb_rejected
[0]
>>> tholmbonf = TestHolmBonferroni(name="holm-bonf",
... description="Holm-Bonferroni method",
... test=tstudent_12, alpha=0.05)
>>> tholmbonf.alpha # alpha is two-sided
0.025
>>> thb_res = tholmbonf.evaluate()
>>> bool(thb_res)
True
>>> thb_res.nb_rejected
[0]
Failing test with Bonferroni correction and Holm-Bonferroni method
>>> ds3 = Dataset(np.array([5.1, 5.9, 5.8, 5.3, 4.6]),
... np.array([0.1, 0.1, 0.05, 0.4, 0.12]))
>>> tstudent_13 = TestStudent(ds1, ds3, name="comp",
... description="Comparison using Student test",
... alpha=0.05, ndf=1000)
>>> tbonf = TestBonferroni(name="bonf", description="Bonferroni correction",
... test=tstudent_13, alpha=0.05)
>>> tbonf.ntests
5
>>> tbonf.bonf_signi_level
0.005
>>> tbonf_res = tbonf.evaluate()
>>> bool(tbonf_res)
False
>>> tbonf_res.nb_rejected
[1]
>>> tbonf_res.rejected_proportion # in percentage
[20.0]
>>> tholmbonf = TestHolmBonferroni(name="holm-bonf",
... description="Holm-Bonferroni method",
... test=tstudent_13, alpha=0.05)
>>> tholmbonf.alpha # alpha is two-sided
0.025
>>> thb_res = tholmbonf.evaluate()
>>> bool(thb_res)
False
>>> thb_res.nb_rejected
[2]
>>> thb_res.rejected_proportion # in percentage
[40.0]
For an easier comparison, p-values, associated significance levels and the corresponding result from the test (rejection or not) can be printed. The sorting order is also given for the Holm-Bonferroni method.
>>> it = np.nditer([thb_res.first_test_res.pvalue,
... tbonf.bonf_signi_level, tbonf_res.rejected_null_hyp,
... thb_res.sort_ordering, thb_res.alphas_i,
... thb_res.rejected_null_hyp],
... flags=['multi_index'])
>>> while not it.finished:
... if it.iterindex == 0:
... print("index | pvalue(B) | α(B) | reject B | order(HB) |"
... f" α(HB) | reject HB\n{'-':->77}")
... print(f"{it.multi_index} | {it[0]:.3e} | {it[1]:.3e} | {it[2]!s:^9} | "
... f"{it[3]:^7} | {it[4]:.3e} | {it[5]!s:<}")
... _ = it.iternext()
index | pvalue(B) | α(B) | reject B | order(HB) | α(HB) | reject HB
-----------------------------------------------------------------------------
(0, 0) | 6.548e-01 | 5.000e-03 | False | 3 | 1.250e-02 | False
(0, 1) | 2.608e-02 | 5.000e-03 | False | 2 | 8.333e-03 | False
(0, 2) | 1.015e-06 | 5.000e-03 | True | 0 | 5.000e-03 | True
(0, 3) | 8.231e-01 | 5.000e-03 | False | 4 | 2.500e-02 | False
(0, 4) | 5.447e-03 | 5.000e-03 | False | 1 | 6.250e-03 | True
The first index corresponds to the index of the compared dataset in the
datasets container in the input test (compared to dsref).
The Holm-Bonferroni test is uniformly more powerful than the Bonferroni test. In this particular case, Holm-Bonferroni is able to reject an additional null hypothesis with respect to plain Bonferroni.
Tests with multi-dimension datasets
>>> ds4 = Dataset(np.array([[5.2, 5.3, 5.25], [5.4, 5.5, 5.2]]),
... np.array([[0.2, 0.25, 0.1], [0.2, 0.3, 0.25]]))
>>> ds5 = Dataset(np.array([[5.1, 5.9, 5.8], [5.3, 4.7, 5.5]]),
... np.array([[0.1, 0.1, 0.05], [0.4, 0.05, 0.2]]))
>>> ds4.shape
(2, 3)
>>> ds4.size
6
>>> tstudent_45 = TestStudent(ds4, ds5, name="comp",
... description="Comparison using Student test",
... alpha=0.05, ndf=1000)
>>> tbonf = TestBonferroni(name="bonf", description="Bonferroni correction",
... test=tstudent_45, alpha=0.05)
>>> tbonf.ntests
6
>>> print(f'{tbonf.bonf_signi_level:.6f}')
0.004167
>>> tbonf_res = tbonf.evaluate()
>>> bool(tbonf_res)
False
>>> tbonf_res.nb_rejected
[1]
>>> print(f'{tbonf_res.rejected_proportion[0]:.3f} %')
16.667 %
>>> tholmbonf = TestHolmBonferroni(name="holm-bonf",
... description="Holm-Bonferroni method",
... test=tstudent_45, alpha=0.05)
>>> thb_res = tholmbonf.evaluate()
>>> tholmbonf.alpha
0.025
>>> bool(thb_res)
False
>>> thb_res.nb_rejected
[1]
>>> print(f'{thb_res.rejected_proportion[0]:.3f} %')
16.667 %
The detailed comparison also works for multi-dimensions arrays:
>>> it = np.nditer([thb_res.first_test_res.pvalue, tbonf.bonf_signi_level,
... tbonf_res.rejected_null_hyp, thb_res.sort_ordering,
... thb_res.alphas_i, thb_res.rejected_null_hyp],
... flags=['multi_index'])
>>> while not it.finished:
... if it.iterindex == 0:
... print("index | pvalue(B) | α(B) | reject B | order(HB) |"
... f" α(HB) | reject HB\n{'-':->79}")
... print(f"{it.multi_index} | {it[0]:.2e} | {it[1]:.2e} | {it[2]!s:^7} "
... f"| {it[3]:^7} | {it[4]:.2e} | {it[5]!s:<}")
... _ = it.iternext()
index | pvalue(B) | α(B) | reject B | order(HB) | α(HB) | reject HB
-------------------------------------------------------------------------------
(0, 0, 0) | 6.55e-01 | 4.17e-03 | False | 4 | 1.25e-02 | False
(0, 0, 1) | 2.61e-02 | 4.17e-03 | False | 2 | 6.25e-03 | False
(0, 0, 2) | 1.01e-06 | 4.17e-03 | True | 0 | 4.17e-03 | True
(0, 1, 0) | 8.23e-01 | 4.17e-03 | False | 5 | 2.50e-02 | False
(0, 1, 1) | 8.66e-03 | 4.17e-03 | False | 1 | 5.00e-03 | False
(0, 1, 2) | 3.49e-01 | 4.17e-03 | False | 3 | 8.33e-03 | False
The first index corresponds to the index of the compared dataset in the
datasets container in the input test (compared to dsref). The next ones
correspond to the shape of the arrays.
The sorting order for Holm-Bonferroni considers all elements as equivalent (not done per dimension).
- class valjean.gavroche.stat_tests.bonferroni.TestResultBonferroni(test, first_test_res, rejected_null_hyp)[source]
Test result from Bonferroni correction.
The test returns
Trueif the test is successful (null hypothesis was accepted) andFalseif it is rejected.- __init__(test, first_test_res, rejected_null_hyp)[source]
Initialisation of
TestResultBonferroni- Parameters:
test (
TestBonferroni) – the used Bonferroni testfirst_test_res (
TestResultchild class) – the test result used to obtain the p-valuesreject_hyp (
numpy.ndarray(bool)) – rejection of the null hypothesis for all bins
- oracles()[source]
Final test (on list of tests, espacially in case of mulitple tests on common reference).
- Returns:
list(bool)
- property nb_rejected
Number of rejected null hypothesis according Holm-Bonferroni test.
- Returns:
list(int) or
list(numpy.generic(int))
- property rejected_proportion
Rejected proportion in percentage.
- Returns:
list(numpy.generic(float))
- class valjean.gavroche.stat_tests.bonferroni.TestBonferroni(*, name, description='', labels=None, test, alpha=0.01)[source]
Bonferroni correction for multiple tests of the same hypothesis.
- __init__(*, name, description='', labels=None, test, alpha=0.01)[source]
Initialisation of
TestBonferroni.- Parameters:
name (str) – local name of the test
description (str) – specific description of the test
labels (dict) – labels to be used for test classification in reports (for example category, input file name, type of result, …)
test (
valjean.gavroche.test.Testchild class) – test from which pvalues will be extractedalpha (float) – required significance level
The significance level is considered two-sided here, so divide by 2. It is the significance level for the whole test.
- property ntests
Returns the number of hypotheses to test, i.e. number of bins here.
- static bonferroni_correction(pvalues, bonf_signi_level)[source]
Bonferroni correction.
- Parameters:
pvalues (
numpy.ndarray(float)) – p-values from the original testbonf_signi_level (float) – significance level for each test (α weighted by the number of tests)
- Returns:
Compares the p-value to the “Bonferroni significance level”:
if lower, null hypothesis is rejected for the bin
if higher, null hypothesis is accepted for the bin.
- class valjean.gavroche.stat_tests.bonferroni.TestResultHolmBonferroni(test, first_test_res, alphas_i, rejected_null_hyp)[source]
Result from the Holm-Bonferroni method.
- __init__(test, first_test_res, alphas_i, rejected_null_hyp)[source]
Initialisation of
TestResultHolmBonferroni- Parameters:
test (
TestHolmBonferroni) – the used Holm-Bonferroni testfirst_test_res (
TestResultchild class) – the test result used to obtain the p-valuessorted_indices (
numpy.generic(int)) – indices of the p-values sorted to get p-values in increasing orderalphas_i (
numpy.ndarray(float)) – significance levels for sorted pvaluesrejected_hyp (
numpy.ndarray(bool)) – rejection of the null hypothesis for all bins
- oracles()[source]
Final test (on list of tests, espacially in case of mulitple tests on common reference).
- Returns:
list(bool)
- property nb_rejected
Number of rejected null hypothesis according Holm-Bonferroni test.
- Returns:
list(int) or
list(numpy.generic(int))
- property rejected_proportion
Rejected proportion in percentage.
- Returns:
list(numpy.generic(float))
- property sort_ordering
Get the sorted pvalues.
- Returns:
list(numpy.generic(int))
- class valjean.gavroche.stat_tests.bonferroni.TestHolmBonferroni(*, name, description='', labels=None, test, alpha=0.01)[source]
Holm-Bonferroni method for multiple tests of the same hypothesis.
- __init__(*, name, description='', labels=None, test, alpha=0.01)[source]
Initialisation of
TestHolmBonferroni.- Parameters:
name (str) – local name of the test
description (str) – specific description of the test
labels (dict) – labels to be used for test classification in reports (for example category, jdd name, type of result, …)
test (
valjean.gavroche.test.Testchild class) – test from which pvalues will be extractedalpha (float) – significance level
The significance level is also considered two-sided here, so divide by 2. This is the overall significance level.
- property ntests
Returns the number of hypotheses to test, i.e. number of bins here.
- static holm_bonferroni_method(pvalues, alpha)[source]
Holm-Bonferroni method.
- Parameters:
pvalues (
numpy.ndarray) – array of pvaluesalpha (float) – significance level chosen for the Holm-Bonferroni method
- Returns:
sorted indices, array of the bins significance level, array of rejection of the null hypothesis